PGAdb-builder User Guide
Step 1: Build PGA Database
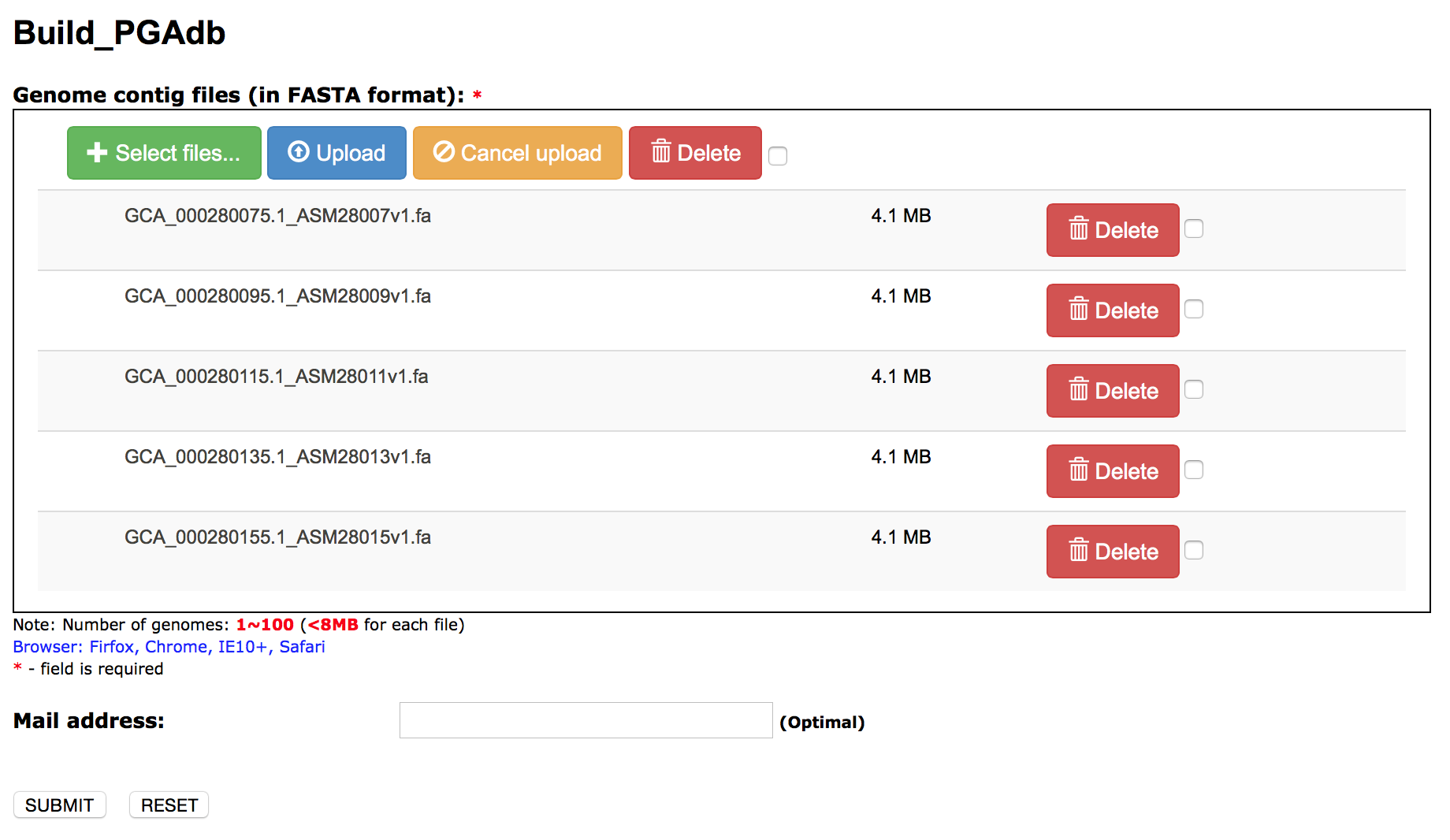
Build_PGAdb input: upload genome contig files
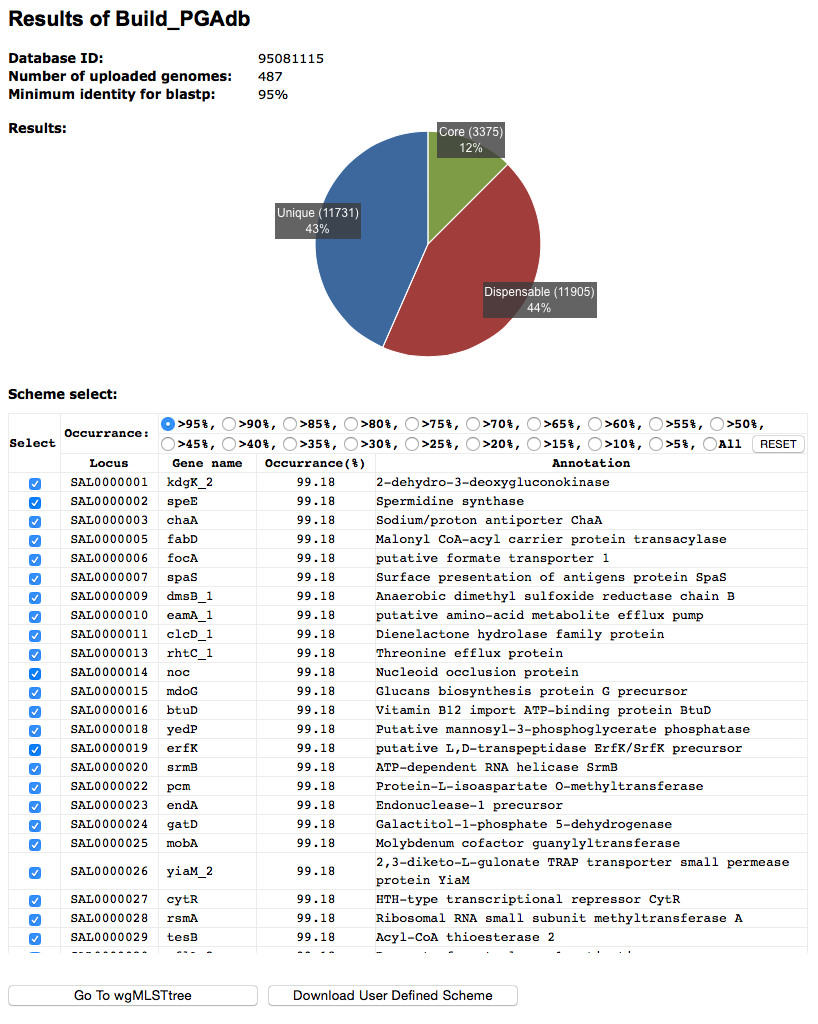
Build_PGAdb output: graphical output example and explanation
The above image is an example of the graphical output of the Build_PGAdb.
Content of the Build_PGAdb output
- A summary of the setting used in the execution
- A summary of output files that users can download
- A pie chart shows the composition of the pan genome database
- A continue execution button for Build_wgMLSTtree
Step 2: Build wgMLST Tree
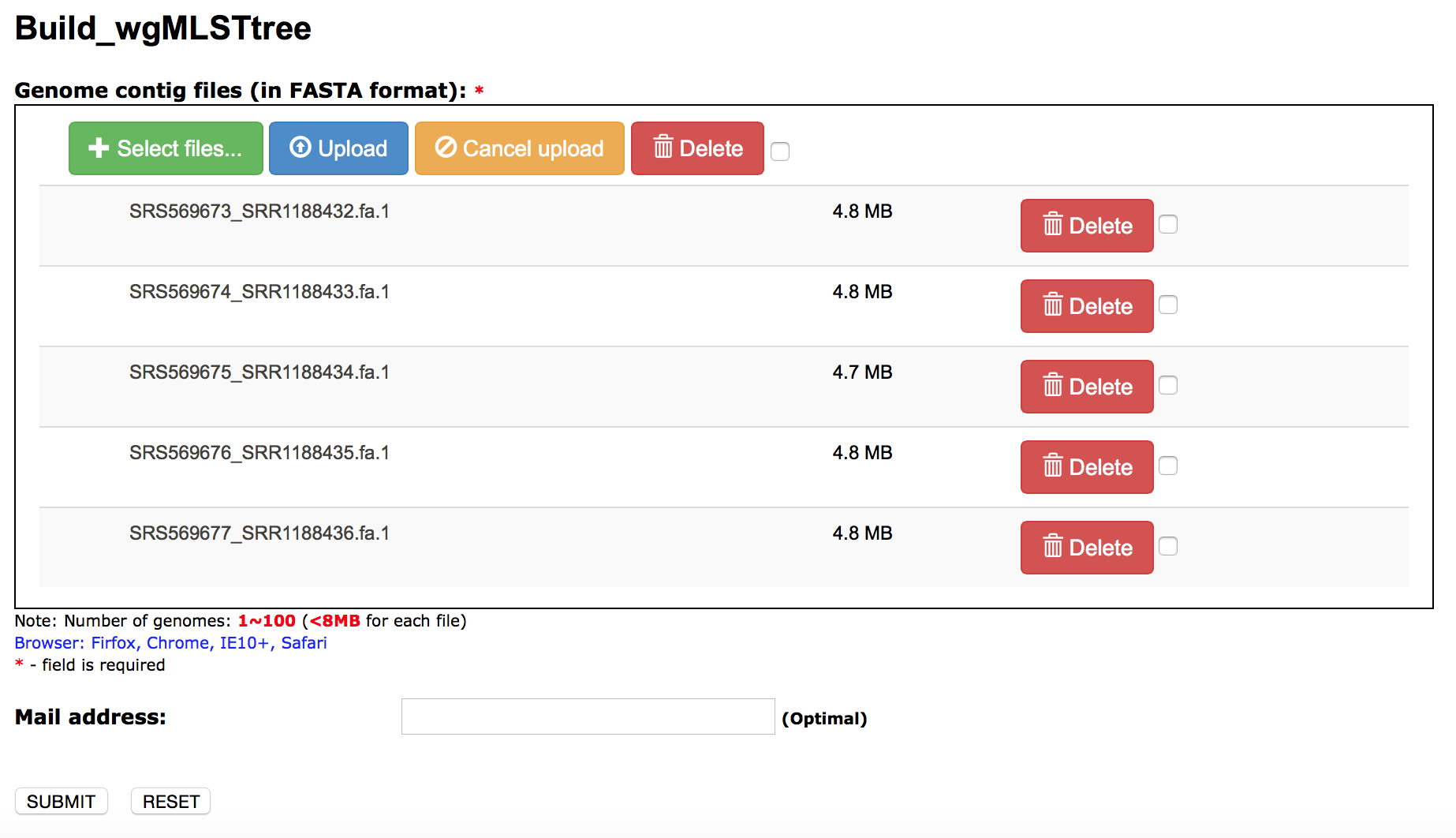
Build_wgMLSTtree input: upload genome contig files
-
User can use their own PGAdb for Build_wgMLSTtree by inputing the unique "Database ID".
-
The parameters define the thresholds of "coverage" and the "identity" when blasting the uploaded genome contigs to the PGAdb.
-
The "loci set" for Build_wgMLSTtree.
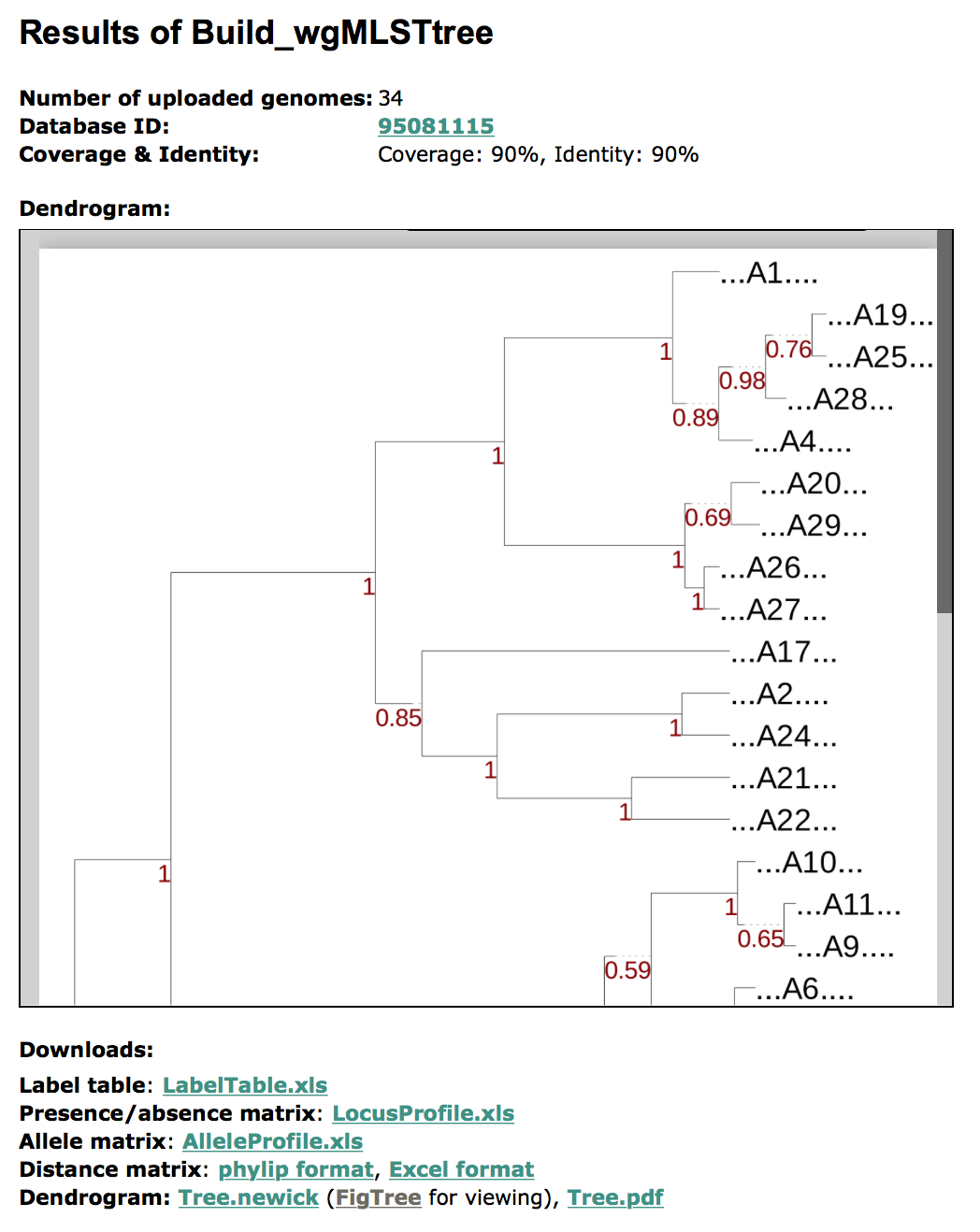
Build_wgMLSTtree output: graphical output example and explanation
The above image is an example of the graphical output of the Build_wgMLSTtree.
Content of the Build_wgMLSTtree output
- A summary of the setting used in the execution
- A summary of output files that users can download
- A execution button for regenerating the genetic relatedness tree
|